 |
Bio-Feedback/Brain Computer Interface

The Bio-Feedback Framework (BF++) has won the "Engineering"
Award at the second Brain Computer Interface Workshop (Albany, NY
12-16 June 2002).
The work that I did and that is described here and in the
related pages is dedicated to Renato Grasso and Alek Kostov
OVERVIEW
The Bio-Feedback SDK (BF++) is a C++ framework that
helps you to develop a Bio-Feedback system, providing an Object Oriented
model that is highly portable under many
Operating Systems and adaptable to a wide range of
situations. It has been designed with an high abstraction level with
respect to the hardware to make it possible (and easy) to use multiple
and mixed input sensors (EMG, breath, voice, digital video, EEG) in
order to use all the patients residual capabilities or to change the
input/output devices without the necessity to rebuild the whole system.
The Bio Feedback SDK (BF++) can be obtained upon request to me,
and it was originally developed for Brain-Computer Interface applications.
BRAIN COMPUTER INTERFACES
Brain Computer Interfaces (BCIs) based on Electroencephalographic
(EEG) signals require on-line detection of mental states
from spontaneous activity: different cortical areas are activated while
thinking different things (i.e. a matematical computation, an imagined
arm movement, a music composition, etc...). The identification of these
different "mental states" can be used to drive external devices:
a mouse cursor can be moved while a subject thinks for example to move
the right arm (i.e. cursor moves into the right direction), the left
arm (left direction), etc... This is a great opportunity for disabled
people with severe impairments.
DOCUMENTATION :-(((
Today, it lacks of a well structured documentation (e.g. help file,
examples, etc..), but it will be available soon (I will release the
first draft in September). The only docs that can give you an idea about
the principles of the proposed model are the following ones:
- the works presented in the 3rd
International Conference of BioElectroMagnetism, in Bled (Slovenia).
To read it click here.
To see the Power Point presentation of the 3rd International Conference
of BioElectroMagnetism, click here
(200KB). To download the zipped version (120 KB) click here.
Note that some improvements have been made since that event;
- A full paper will be published in 2003 about BF++ in "Computher
Methods of Information in Medicine". It was originally developed
to build a Brain Computer Interface system;
- A short paper has also been submitted and it will be hopefully be
published in early 2003.
- I did another "quick" PowerPoint presentation presented
before the wonderful talk of Prof. Jonathan
Wolpaw at St. Lucia Rehabilitation Hospital (Rome, Italy),
the 15th of December 2000. You can see my presentation (without videos
and ... voice!) here.
The presence of such a BCI Guru has been a great opportunity to learn
a lot of things from a scientist that has done the history and is
doing the... future of Brain Computer Interface. The event was completed
with the great presentation held by our "friend" Josè
Millan. I'll remember that day forever!
ACKNOWLEDGMENTS
While I did the dirty work of writing the majority of the code, this
SDK is the result of the work, suggestion and feedback of a research
group, and that without them...
The Team is actually formed by:
Prof. Maria Grazia Marciani, that is my Chief and that for
this reason is the best person in all over the world!
Prof. Fabio Babiloni, that is the Master and that has
"just" written some important pages in EEG literature.
Febo Cincotti, that was the "inventor" of the italian
BCI connection (and one of the most acute researchers I've ever known).
Also in the past worked at the italian BCI connection:
Francesco Semeraro, engineer,
Angela Cattini, engineer,
Vittorio Gigli, engineer.
I also would like to thank, for their support, inspiration and patience,
my wife, Luisa, and my daughter Ludovica,
that accepted that I did this work for free, so that they had a "part-time"
husband/father for long "time intervals". In particular I would
also like to thank the Wadsworth Center, for being my main scientific
sponsors and for giving me the main opportunities to share my ideas with
the community. For different reasons I have to thank: Jonathan Wolpaw
and his wonderful wife Liz, Theresa Vaughan, Gervin Schalk, Dennis McFarland
and all the other memebers of the team. I also would like to thank Niels
Birbaumer, Alan Gevins, Fernando Lopez da Silva, Andrea Kuebler, nd all
the friends of mine that encouraged me in this reasearch field. Finally,
I have also to mention the EBNeuro staff that provided me some very useful
prototypes and revealed me many industrial secrets.
Some more (confusing) info about BF++
A set of C++ classes have been defined in order to easily develop a
Bio-Feedback system. They have been designed with the aim of being independent
on the platform used (both SW and HW) because a main goal is to use
it with a Palm PC or in an embedded system. Also code reuse was
a main goal, because it is fundamental for disables to have a wide range
of opportunities to use every residual
capability that they have.
Matrix computation and FFT routines are also included. Actually the
Signal Space Projection implementation
requires about 50 lines of code.
Implementing a solution based on Mahalanobis distance required also
less than 60 lines of code.
Actually the system runs on a Windows machine, on Linux (demonstration
and off-line analisys) and Windows CE (there is a working BCI system
prototype), but this is not required. Anyway, even if
the framework is platform independent, one can use some features that
are Win32 specific: in particular, in a BCI application of mine, the
3-D Spectral Maps (computed in "realtime") are based on Microsoft
Direct3D technology, while voice commands and speech synthesis
are obtained through the MS Speech API (4.0) and IBM ViaVoice SDK (for
the italian voice engine). I'm also working in the integration of Accessibility
options provided by Windows (see the
MSDN Magazine publication).
Status:
After the BCII meeting (Albany, 12-16 June 2002) I am rewriting some
minor things and a complete documentation.
The programming books that influenced the new release are:
Modern C++ Design: Generic Programming and Design
Patterns Applied (Andrei Alexandrescu, Addison Wesley 2001)
Design Patterns CD: Elements of Reusable Object-Oriented
Software (Erich Gamma et al., Addison Wesley, 1998)
Effective C++ CD: 85 Specific Ways to Improve
Your Programs and Designs (Scott Meyers, Addison Wesley, 1999)
Exceptional C++: 47 Engineering Puzzles, Programming
Problems, and Solutions (Herb Sutter, Addison Wesley, 2000)
The C++ Standard Library: A Tutorial and Reference
(Nicolai Josuttis, Addison Wesley, 2000)
The framework is described by the following UML chart (click to enlarge):
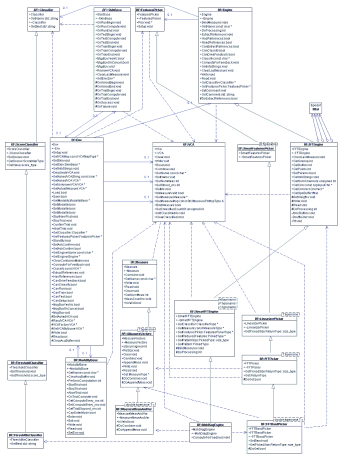 |
BF::Env
is the concertmaster of the framework. It holds a reference to a
BF::Engine, which is
responsible of the "algorithmic" aspects, some BF::VCAs
which describe the volountarily controlled activity that the subject
wants to use (e.g. the imagination of a movement), and a BF::Skin
which provides the feedback to the user. To each VCA one or more
BF::Measures
can be associated. At this level, it is not important to define
what these measures do represent: they could be mean values,
images, covariance matrices or a set of them. It is sufficient to
have the opportunity to bind one or more generic measures to each
VCA. This is important beacause usually BF systems implement a training
phase and a running phase. During the training the system have to
extract some reference measures that are later on used in the running
modality: acquired data are compared with the reference measures
and then a classification is demanded to the execution of a "best
match" routine. This last task is performed by the BF::Classifier.
Sometimes a subset of a measure is needed (e.g. if the measure is
a spectrum usually a limited spectral band is sufficient) and is
the task performed by the BF::FeaturesPicker
object.
To implement a BF system it is sufficient to inherit some of the
provided helper classes and customize their behaviour. For example,
the SSP (Signal Space Projection) implementation uses other helper
objects that are provided to the framework: the FFTEngine
performs the spectral analysis of some temporal series, while the
SmartFFTEngine provides (through templates)
some more customization options that are finally specialized in
the SSPEngine. It has to override just two
virtual functions: DoExtractPattern that
is invoked when the system switches from the training to the running
mode and ClassRule that is invoked whenever
a classification is tried. In the case of the SSP system that's
all, because other helper classes are used to get the mean spectra
of the input signals while the FeaturePicker
(and descendents) automatically select just a part of the mean spectra
data (e.g. a spectral band). Put in other words: the "SSP system"
has been implemented by specializing a SmartFFTEngine (using a MeasureMean
object, a ThresholdMax object, and FFTBandPicker
object) and overriding two virtual functions. |
This is the result:
//begin of code
#include "stl.h"
#include <fstream>
#pragma hdrstop
#include "SSPEngine.h"
#include "matrix_utils.h"
#include "newmatap.h"
SSPEngine::SSPEngine(ChnVect *chnv, int _fft_points, int _fft_buff_count, int _fft_overlap) :
SmartFFTEngine<ThresholdMax, MeasureMean<>, FFTBandPicker<> >
(chnv, _fft_points, _fft_buff_count, _fft_overlap)
{ GetFeaturesPicker()->SetBand(8., 30.); }
bool SSPEngine::DoExtractPatterns(const VCAMap& sm)
{ Inherited::DoExtractPatterns(sm, rest_state);
NEWMAT::Matrix pseudo_inv(sm.size(), GetPatternSize(sm.begin()->second));
fPseudoInv.ReSize(pseudo_inv.Nrows(), pseudo_inv.Ncols());
NEWMAT::Matrix _mat(pseudo_inv.Ncols(), pseudo_inv.Nrows());
PickedType meas;
for (vca_cIt i = sm.begin(); i != sm.end(); ++i)
{ meas = GetPattern((*i).second, "Mean");
for (unsigned int index = 0; index < meas.size(); ++index)
_mat(index + 1, distance(sm.begin(), i) + 1) = meas[index];
}
NEWMAT::DiagonalMatrix D;
NEWMAT::Matrix U, V;
SVD(_mat, D, U, V);
pseudo_inv = V * D.i() * U.t();
for (unsigned int r = 0; r < fPseudoInv.Nrows(); r++)
for (unsigned int c = 0; c < fPseudoInv.Ncols(); c++)
fPseudoInv[r][c] = pseudo_inv[r][c];
return true;
}
void BF_SSPEngine::ClassRule(const VCAMap& sm, VCA* am_)
{ fMat matrixState(fPseudoInv * fMat(GetPattern(am_, "Mean")));
ScoreMapType smt;
for (vca_cIt i = sm.begin(); i != sm.end(); ++i)
smt[(*i).first] = matrixState[distance(sm.begin(), i)][0];
GetClassifier()->SetScores(smt);
}
//end of code
While I do not pretend that you understand from these few (uncommented!)
lines how the framework works (an article will be published in "Computer
Methods of Information in Medicine"), it is important that you
realize that with just few lines of code you can build a whole working
BF system, and that the code is portable under every platform that supports
a C++ compiler. What you have to do is simply override few virtual functions,
that usually are: DoExtractPatterns,
which is invoked when the training is finished and the user switches
to the running modality, DoProcessing
which is often already available as it is provided by an existing engine
and that is responsible of the computation, and ClassRule,
which is responsible in the running modality to compare the processed
data with the Reference Pattern data (fPseudoInv).
Just two more words about the constructor. The following lines are
pure dynamite:
SSPEngine::SSPEngine(ChnVect
*chnv, int _fft_points, int _fft_buff_count, int
_fft_overlap) :
SmartFFTEngine<ThresholdMax,
MeasureMean<>,
FFTBandPicker<> >
(chnv, _fft_points, _fft_buff_count, _fft_overlap)
{ GetFeaturesPicker()->SetBand(8., 30.);
}
What they do is great: SSPEngine is an engine with FFT capabilities.
Each time that a measure is performed the MeanSpectra of the input signals
are computed automagically as we have decided it (see
2nd template parameter MeasureMean<>). Optionally we could
choose to compute the Mean and SD by simply replacing MeasureMean with
MeasureMeanAndSD<>. The FFTBandPicker is another
object that allows us to use just an FFT Band for all the computations
(probably frequencies above 40 Hz are useless in EEG based systems).
This is the Feature reduction policy: if we want to use only the data
whose amplitude exceeds a certain value it is sufficient to provide
a valid FeatureReduction derived class. Finally, ThresholdMax
means that a classification occurs in the following way: to each VCA
a floating point value is associated and if the Max value exceeds the
second to the best by at least a given Threshold then a classification
occurs, otherwise the trial remains unclassified. Now, hopefully, it's
less obscure how it is possible to build a BCI system with just few
lines of code.
Some Specs about
a Win32 implementation (ver. 1.1) compiled with the free bcc55 Borland
command line compiler - except the SkinWin1.DLL which uses Borland C++
Builder 5 (BCB5), with the Embedded Visual C++ 3.0 (EVC 3, X86, ARM
and MIPS processors), with Microsoft VC 6.0 (VC6), with Microsoft VC
7.0 (VC7) with and without managed extensions (.NET platform) and the
free gcc 2.95.3-6:
| DLL |
VC6 |
VC7 (unma-naged) |
VC7 (mana-ged) |
EVC 3.0 x86 |
EVC 3.0 ARM
|
EVC 3.0 MIPS |
GNU GCC 2.95.3-6
|
BCB5 |
Functionalities |
| BFKernel.dll |
64 KB |
64 KB |
176 KB |
45 KB |
68 KB |
92 KB |
154 KB |
94 KB |
This is the kernel of the Bio-Feedback SDK.
It includes the basics for every BF engine. Effort has been taken
to minimize its size. |
| XMLUtils.dll |
72 KB |
68 KB |
156 KB |
--- |
--- |
--- |
161 KB |
91 KB |
A DLL generated from the source code of Sebastien
Andrivet (AdvXMLParser
Version 1.1.4c) and which is used for loading and saving the
files. |
| Newmat.dll |
208 KB |
196 KB |
344 KB |
150 KB |
218 KB |
249 KB |
353 KB |
271 KB |
This the cross-platform Matrix C++ framework
developed by Robert Davies (called NEWMAT). Latest
updates can be found here.
It is not required to build a Bio-Feedback system. It is, however,
the best C++ class library I've found on the net. And it's free!
BTW, it is used by the Signal Space Projection (SSP) algorithm. |
| BCIStub.dll |
64 KB |
68 KB |
156 KB |
28 KB |
38 KB |
41 KB |
133 KB |
80 KB |
A Win32 GUI BCI customizer: it allows to select the
engine and skin to use and allows also to set a lot of parameters. |
| FFTEng.dll |
32 KB |
22 KB |
60 KB |
12 KB |
17 KB |
21 KB |
114 KB |
36 KB |
A generic engine which add FFT capabilities. |
| Win32skn.dll |
24 KB |
12 KB |
22 KB |
5 KB |
6 KB |
7 KB |
94 KB |
22 KB |
A generic (abstract) skin for Win32 applications.
It implements some GUI aspects such as MessageBox, etc.. |
| Win32SDK.skn |
48 KB |
34 KB |
82 KB |
20 KB |
25 KB |
28 KB |
139 KB |
62 KB |
A skin entirely written using the plain SDK. |
| SkinWin1.skn |
--- |
--- |
--- |
--- |
--- |
--- |
--- |
132 KB + VCL |
This is a Win32 specific skin for linear classifiers.
It still depends on VCL which makes it compile just with Borland
C++ Builder 5. Actually I'm solving this problem. This is the also
only DLL which contains code that is specific to Microsoft Windows.
The other ones can be recompiled under different platforms/C++ compilers. |
| MahDiag.eng |
40 KB |
44 KB |
108 KB |
19 KB |
29 KB |
41 KB |
125 KB |
60 KB |
This is an implementation of the Mahalanobis Diagonal
algorithm for a BCI system. This is also platform independent. |
| SSP.eng |
--- |
48 KB |
120 KB |
--- |
--- |
--- |
140 KB |
74 KB |
This is an implementation of the SSP algorithm for
a BCI system. This is also platform independent. |
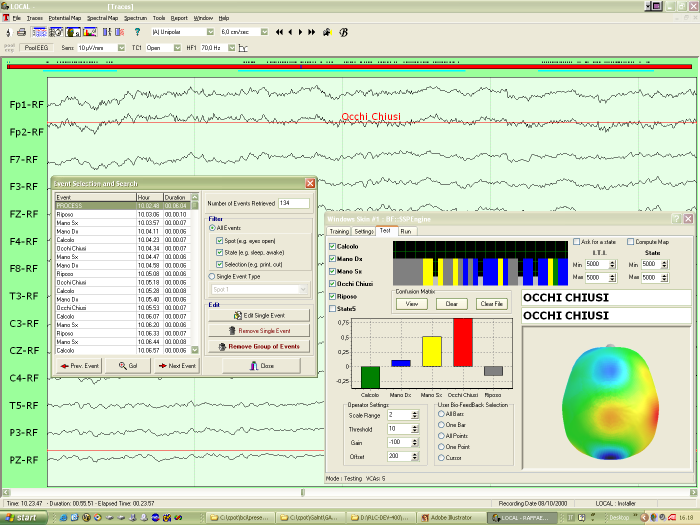
Here you can have a screenshot of a Brain Computer Interface application:
the SSP engine has been plugged-in in a commercial EEG system (Galileo,
by EBNeuro).
Related Publications
Bianchi L, Babiloni F, Cincotti F, Salinari
S, Marciani MG, Introducing BF++: A C++ Framework for Cognitive Bio-Feedback
Systems Design, Methods of Information in Medicine, in press
Febo Cincotti, Claudio Babiloni, Filippo
Carducci, Luigi Bianchi, José del R. Millán, Josep Mouriño,
Serenella Salinari, Maria Grazia Marciani and Fabio Babiloni (2002),
Classification of EEG mental patterns by using two scalp electrodes
and Mahalanobis distance-based classifiers, Methods of Information in
Medicine, December 2002
Babiloni F, Cincotti F, Bianchi L, Pirri G,
Millán J, Mouriño J, Salinari S, and Marciani MG (2001),
Recognition of Imagined Hand Movements with Low Resolution Surface Laplacian
and Linear Classifiers, Medical Engineering & Physics, Vol 23/5,
pp 323-328
Babiloni F, Cincotti F, Bianchi L, Millán
J, Mouriño J, Salinari S, Marciani MG (2001). Recognition of
Imagined Hand Movements with Low Resolution Surface Laplacian and Linear
Classifiers, Human Brain Mapping 2001, 10-14 June, Brighton, U.K.
Bianchi
L, Babiloni F, Cincotti F, Salinari S, Marciani MG (2000). An Object
Oriented Approach To Biofeedback Applications For Disabled People, 3
International Conference on BioElectroMagnetism, Bled, Slovenia, October
2000.
Babiloni F, Cincotti F, Bianchi L,
Pirri G, Millán J, Mouriño J, Salinari S, Marciani MG (2000). Recognition
of Imagined Hand Movements by Signal Space Projection and Fisher Discriminant
Linear Classifiers, 3 International Conference on BioElectroMagnetism,
Bled, Slovenia, October 2000.
Cincotti F, Mouriño J, Pirri G, Millán
J, Bianchi L, Salinari S, Marciani MG, and Babiloni F (2000). Low Resolution
Surface Laplacian Computation for Brain Computer Interface Classifiers,
3 International Conference on BioElectroMagnetism, Bled, Slovenia, October
2000.
Babiloni F, Cincotti F, Lazzarini L,
Millan J, Mouriño J, Varsta M, Heikkonen J, Bianchi L, Marciani MG.
(2000). Linear Classification of Low-Resolution EEG Patterns produced
by Imagined Hand Movements. IEEE Trans. On Rehabil. Engng., Piscataway,
New Jersey, USA, June, 2000.
Babiloni F, Cincotti F, Bianchi L,
Lazzarini L, Millán J, Mouriño J, and Marciani MG (1999). Surface Laplacian
Electrodes for Adaptive Brain Interfaces, First Brain Computer Interface
World Conference, Albany, July 1999, pag. 40–43.
Babiloni F, Cincotti F, Bianchi L,
Lazzarini L, Millán J, Mouriño J, and Marciani MG (1999). On the Use
of Laplacian Electrodes for Adaptive Brain Interfaces. 3rd International
Workshop on Biosignal Interpretation, pp. 354–357. June 12-14, 1999,
Chicago, USA.
Babiloni F, Cincotti F, Lazzarini L,
Millan J, Mouriño J, Varsta M, Heikkonen J, Bianchi L, Marciani MG
(1999). EEG recognition of imagined hand and foot movements through
signal space projection. Medical & Biological Engineering &
Computing, London, UK ,37(2): 542-543, 1999.
© Copyright (2000), Luigi Bianchi
Last Update:
February 14, 2003
|



